Visualization
Info
If you want to run the code below, consider downloading the demo data or use the TNGLab online.
Creating plots
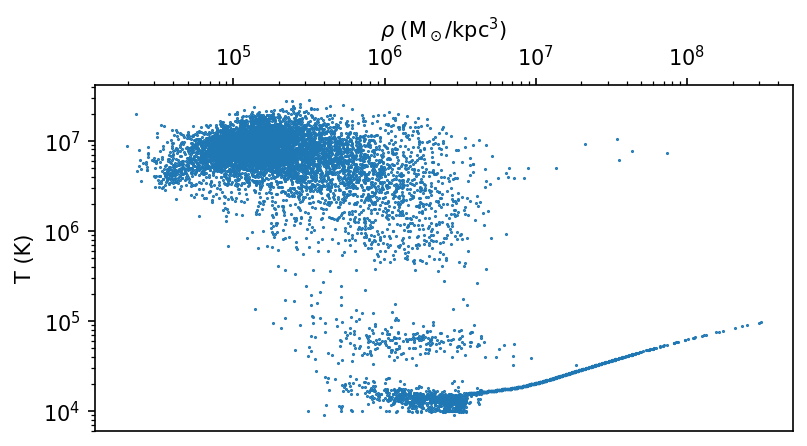
As we often use large datasets, we need to be careful with the amount of data we plot. Generally, we reduce the data by either selecting a subset or reducing it prior to plotting. For example, we can select a subset of particles by applying a cut on a given field.
from scida import load
import matplotlib.pyplot as plt
ds = load("./snapdir_030")
dens = ds.data["PartType0"]["Density"][:10000].to("Msun/kpc^3").magnitude.compute() # (1)!
temp = ds.data["PartType0"]["Temperature"][:10000].to("K").magnitude.compute()
fig, ax = plt.subplots(figsize=(6, 3))
ax.plot(dens, temp, "o", markersize=0.5)
ax.set_xscale("log")
ax.set_yscale("log")
ax.set_xlabel(r"$\rho$ (M$_\odot$/kpc$^3$))")
ax.set_ylabel(r"T (K)")
ax.xaxis.tick_top()
ax.xaxis.set_label_position('top')
plt.show()
- Note the subselection of the first 10000 particles and conversion to a numpy array. Replace this operation with a meaninguful selection operation (e.g. a certain spatial region selection).
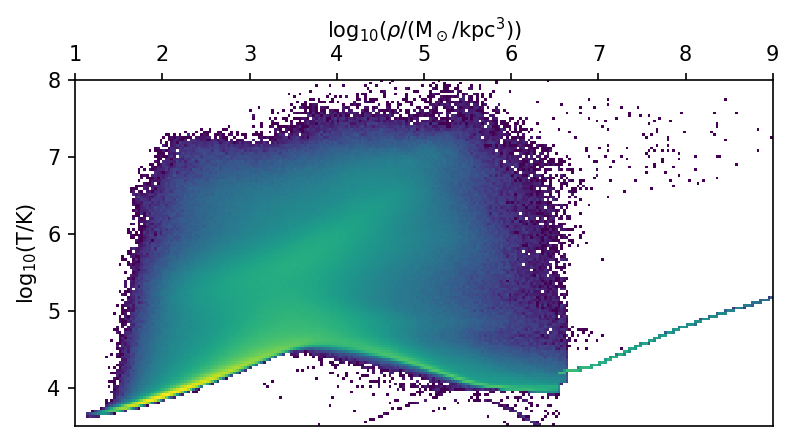
Instead of subselection, we sometimes want to visualize all of the data. We can do so by first applying reduction operations using dask. A common example would be a 2D histogram.
import dask.array as da
import numpy as np
from scida import load
import matplotlib.pyplot as plt
from matplotlib.colors import LogNorm
ds = load("./snapdir_030")
dens10 = da.log10(ds.data["PartType0"]["Density"].to("Msun/kpc^3").magnitude)
temp10 = da.log10(ds.data["PartType0"]["Temperature"].to("K").magnitude)
bins = [np.linspace(1, 9, 256 + 1), np.linspace(3.5, 8, 128 + 1)]
hist, xedges, yedges = da.histogram2d(dens10, temp10, bins=bins)
hist, xedges, yedges = hist.compute(), xedges.compute(), yedges.compute()
extent = [xedges[0], xedges[-1], yedges[0], yedges[-1]]
fig, ax = plt.subplots(figsize=(6,3))
ax.imshow(hist.T, origin="lower", extent=extent, aspect="auto", cmap="viridis",
norm=LogNorm(), interpolation="none")
ax.set_xlabel(r"$\log_{10}$($\rho$/(M$_\odot$/kpc$^3$))")
ax.set_ylabel(r"$\log_{10}$(T/K)")
ax.xaxis.tick_top()
ax.xaxis.set_label_position('top')
plt.show()
Interactive visualization
Info
This example requires the holoviews, datashader and bokeh packages installed.
Make sure that these holoviews examples work before continuing.
The conversion from hdf5-backed dask arrays to dataframes seems to be broken with recent dask versions, this example might currently not work.
We can do interactive visualization with holoviews. For example, we can create a scatter plot of the particle positions.
import holoviews as hv
import holoviews.operation.datashader as hd
import datashader as dshdr
from scida import load
ds = load("./snapdir_030")
ddf = ds.data["PartType0"].get_dataframe(["Coordinates0", "Coordinates1", "Masses"]) # (1)!
hv.extension("bokeh")
shaded = hd.datashade(hv.Points(ddf, ["Coordinates0", "Coordinates1"]), cmap="viridis", interpolation="linear",
aggregator=dshdr.sum("Masses"), x_sampling=5, y_sampling=5)
hd.dynspread(shaded, threshold=0.9, max_px=50).opts(bgcolor="black", xaxis=None, yaxis=None, width=500, height=500)
- Visualization operations in holowview primarily run with dataframes, which we thus need to create using this wrapper for given fields.